How to Build No-Code AI Workflows for Bioinformatics (Simple Guide 2026)

Create powerful bioinformatics analysis without writing a single line of code
Why No-Code Matters in Bioinformatics
Bioinformatics used to be only for people who could code. If you wanted to analyze DNA sequences or study gene expression, you needed to learn Python or R. This limitation prevented many talented biologists from conducting their own computational analyses. They understood biology deeply but could not write the code to answer their research questions.
Today, things have changed completely. No-code platforms let you build complex bioinformatics workflows using simple drag-and-drop interfaces. You click, connect, and configure instead of coding. Even better, artificial intelligence now helps you build these workflows by understanding what you want in plain English.
This guide shows you exactly how to create your first no-code bioinformatics workflow. Whether you are a biologist who wants to analyze your own data, a student learning bioinformatics, or a researcher exploring new methods, you will learn practical steps to get started. No programming experience needed.
What Is a No-Code AI Workflow
Understanding No-Code Workflows
A no-code workflow is like building with blocks. Each block represents a tool or analysis step. You connect these blocks to create a complete analysis pipeline. For example, one block might clean your data, another aligns DNA sequences, and a third creates visualizations. You arrange and connect them visually, rather than writing code.
Think of it like making a flowchart. You draw boxes for different steps and arrows showing how data moves from one step to the next. The platform handles all the technical details behind the scenes. You focus on the science while the software manages the coding.
How AI Makes It Even Easier
Modern platforms add artificial intelligence to help you build workflows faster. You can describe what you want in normal language, like "I want to find differentially expressed genes in my RNA-seq data." The AI suggests which tools to use, how to connect them, and what settings work best.
AI also checks your workflow for mistakes before you run it. It warns you if something looks wrong and suggests fixes. Some platforms even write custom code for you when you need something special that standard tools cannot do.
Why This Matters for Your Research
No-code workflows save time and reduce errors. What used to take weeks of coding can now be done in hours. You avoid common programming mistakes that waste days of troubleshooting. Most importantly, you spend more time understanding your biological results instead of fighting with code.
These platforms also help teams work together better. A computational expert can build a workflow template that everyone in the lab uses. This ensures everyone analyzes data the same way, making results more consistent and reliable.
Step by Step: Build Your First Workflow
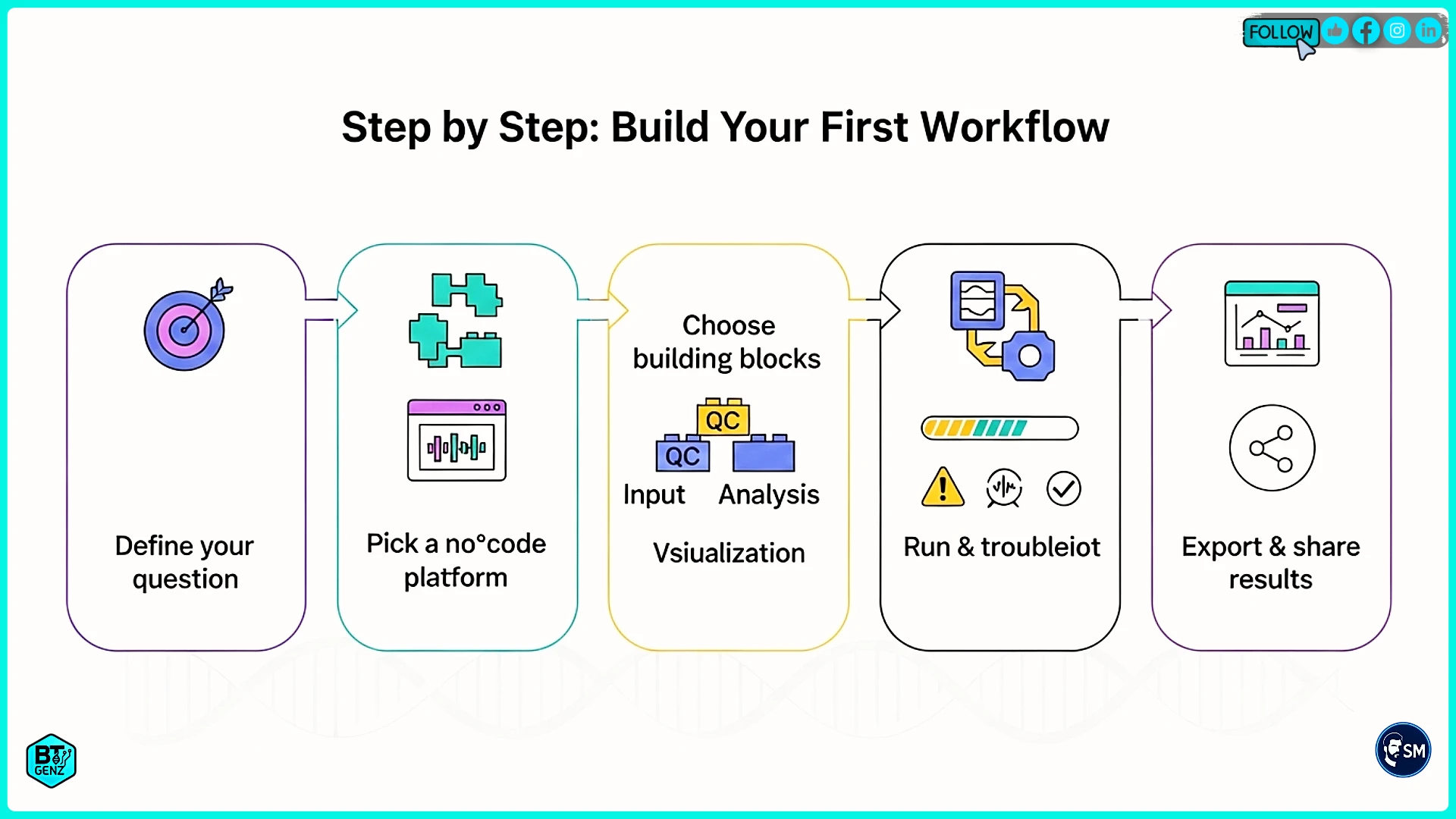
Step 1: Define Your Goal
Start by writing down exactly what you want to learn from your data. Be specific. Instead of "analyze my sequencing data," write "identify which genes are expressed differently between cancer cells and normal cells." Clear goals lead to better workflows.
Write down these details about your project. What type of data do you have? How many samples? What comparisons do you want to make? What biological question are you trying to answer? This information helps you choose the right tools and build an appropriate workflow.
Step 2: Choose Your Platform
Several platforms let you build no-code bioinformatics workflows. Each has different strengths. Here are the main options to consider.
Galaxy is the most popular free platform with thousands of tools available. It works well for standard analyses like RNA-seq, variant calling, and genome assembly. Being free and open source makes it perfect for students and academic researchers. The interface takes some learning but has excellent tutorials.
GenePattern offers a cleaner, simpler interface with fewer but well-documented tools. Developed by the Broad Institute, it focuses on genomics and gene expression analysis. Great for beginners because it guides you through common analyses step by step.
Bionl represents the newest generation with built-in AI. You can describe what you want in plain English, and it builds workflows for you. The AI also writes custom code when needed. Perfect if you feel uncertain about which tools to use or how to connect them.
DNAnexus is a commercial platform used by hospitals and pharmaceutical companies. It costs money but provides a secure, compliant infrastructure for clinical genomics. Choose this if you work with patient data requiring strict privacy controls.
For learning, start with Galaxy or GenePattern. Both are free and have excellent learning resources. Try Bionl if you want maximum AI assistance. Move to commercial platforms later if your work requires their special features.
Step 3: Prepare Your Data
Good data preparation prevents most workflow problems. Organize your files clearly with names that tell you what each file contains. Create a spreadsheet documenting your samples, what group each belongs to, and any important details about how you collected the data.
Check that your data files are in the correct format. DNA sequencing data usually comes as FASTQ files. Gene expression data might be in CSV format. Different tools expect specific formats, so verify yours match what the platform needs.
Before uploading large files, test a small sample first. This catches problems early when fixing them is quick and easy. Once your test works, upload your complete dataset, knowing it will work correctly.
Step 4: Build Your Workflow
Now comes the fun part. Open your chosen platform and look for the workflow builder or editor. Most platforms show a blank canvas where you will add your analysis steps.
Search the tool library for what you need. For RNA-seq analysis, you might search for alignment tools like STAR, counting tools like HTSeq, and statistical tools like DESeq2. Add each tool to your canvas by clicking or dragging it.
Connect the tools by drawing lines between them. Output from one tool becomes input for the next tool. For example, raw sequencing data goes into the alignment tool. Aligned data goes into the counting tool. Count data goes into the statistics tool. The platform usually shows you which connections are valid.
Configure each tool by clicking on it and setting parameters. Most tools have default settings that work fine for common cases. Read the help text if you are unsure what a setting means. Many platforms suggest recommended values for typical data types.
Look for pre-built workflow templates if available. Many platforms have libraries of workflows that other people have shared. Starting from a template is faster and teaches you how experienced users structure their workflows. You can customize the template for your specific needs.
Step 5: Add AI Features
If your platform has AI capabilities, use them to improve your workflow. In platforms like Bionl, you can describe your analysis goal in normal sentences. The AI will suggest a complete workflow, including which tools to use and how to connect them.
AI can also optimize your parameter settings. Instead of guessing which values to use, let the AI analyze your data characteristics and suggest optimal settings. This helps avoid common mistakes that hurt the quality.
Some platforms offer AI debugging. If your workflow fails, the AI reads the error message and explains what went wrong in simple language. It suggests specific fixes instead of showing cryptic technical errors.
Step 6: Run and Check Results
Before running your workflow on all data, test it on one or two samples. This test run catches configuration mistakes quickly without wasting time on large datasets. If the test works, run the complete analysis confidently.
Start your workflow by clicking the run button. Most platforms show progress as it runs. You can see which steps have completed and which are still running. Large datasets take longer, sometimes hours or days, depending on analysis complexity.
When finished, carefully check the results. Look at quality control metrics to ensure the analysis worked correctly. Create plots to visualize patterns in your data. Compare the results to what you expected based on your biological knowledge. Unexpected results might indicate problems or interesting discoveries.
Popular No-Code Tools You Should Know
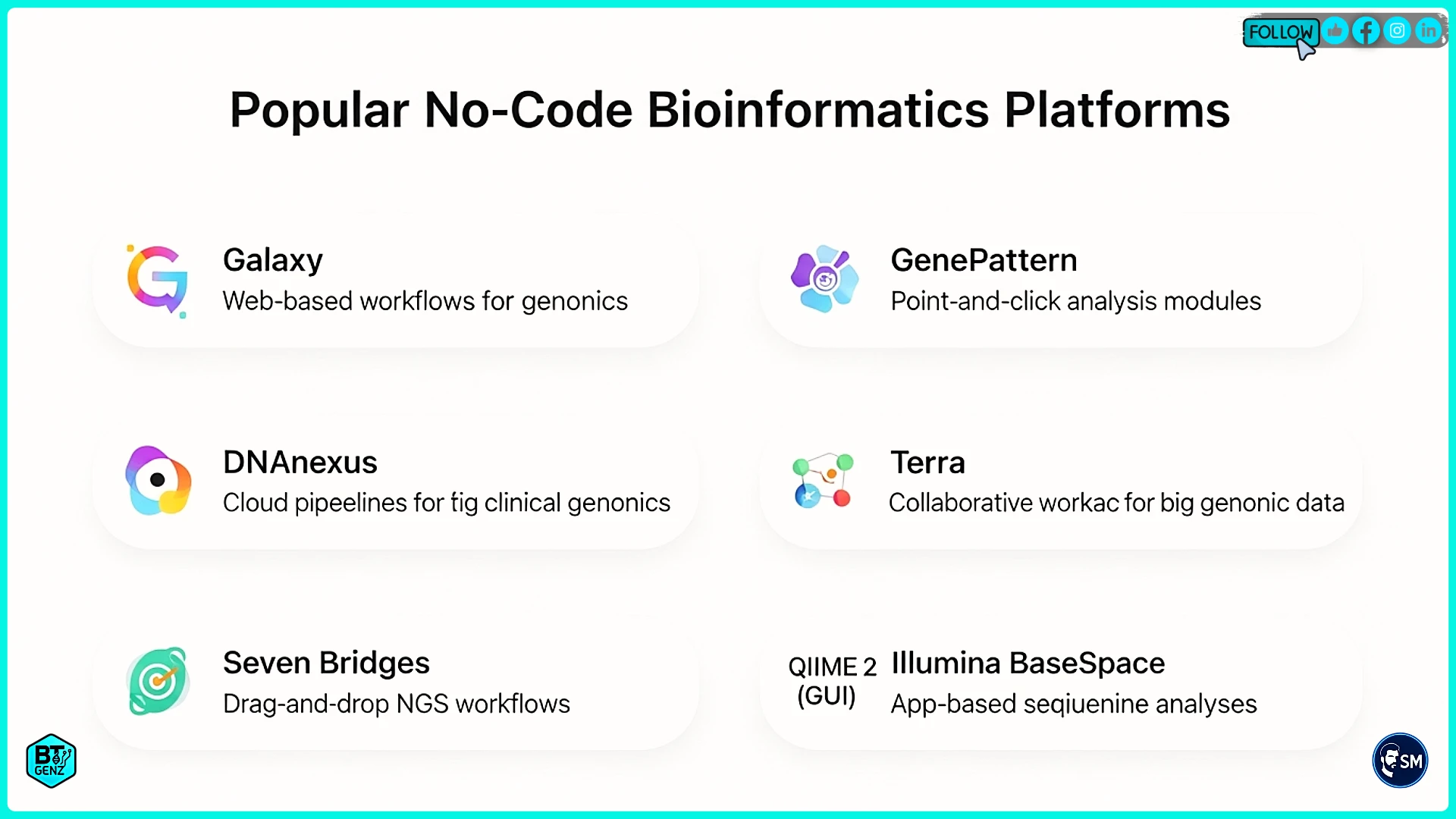
Galaxy for General Bioinformatics
Galaxy dominates academic bioinformatics with over 8,000 tools covering almost any analysis you can imagine. Its visual workflow builder lets you connect tools using drag and drop. Public Galaxy servers provide free access, though you can also install it on your own computers.
Galaxy excels at standard genomics workflows. RNA-seq, variant calling, ChIP-seq, and genome assembly all work excellently. The large tool collection means you rarely need custom code. Strong documentation and tutorials help you learn, though the interface looks older than newer platforms.
GenePattern for Guided Analysis
GenePattern from the Broad Institute focuses on user-friendly interfaces and excellent documentation. It has fewer tools than Galaxy, but each is well explained with examples. This makes it perfect for beginners who feel overwhelmed by too many choices.
The platform specializes in gene expression analysis, genomics, and pathway analysis. Each tool includes detailed help explaining what it does and when to use it. Step-by-step workflows guide you through common analyses from start to finish.
Bionl for AI-Powered Workflows
Bionl represents the future with artificial intelligence built into everything. Describe your analysis in plain English and watch the AI build a workflow for you. It selects appropriate tools, suggests parameter values, and explains each step in simple language.
The AI also generates custom code when you need analysis steps that standard tools cannot provide. This bridges the gap between no-code simplicity and coding flexibility. As a newer platform, Bionl has fewer tools than Galaxy, but its AI capabilities often compensate.
DNAnexus for Clinical Work
DNAnexus serves hospitals and pharmaceutical companies needing secure, compliant infrastructure. It meets the strict privacy regulations required for patient data. The platform scales automatically from small datasets to thousands of genomes.
While commercial and more expensive, DNAnexus provides enterprise features like detailed audit trails, validated workflows for clinical testing, and integration with hospital systems. Choose this if your work involves patient care or regulatory requirements.
Tips for Success
Start Small and Test Often
Always test workflows on small datasets before running complete analyses. Use one or two samples to verify everything works correctly. Testing catches mistakes early, and fixing them is quick. Once your test succeeds, run the full dataset, knowing it will work.
Build workflows step by step rather than creating everything at once. Add one or two tools, test them, then add more. This incremental approach makes debugging easier because you know exactly what changed when something breaks.
Use Templates and Examples
Most platforms offer libraries of shared workflows. Browse these templates for analyses similar to yours. Starting from a working template is much faster than building from scratch. Modify the template for your specific data and questions.
Templates also teach best practices. Experienced bioinformaticians built them, so they show you proper tool selection, parameter settings, and workflow structure. Learn from these examples to improve your own workflow skills.
Document Everything
Write notes explaining why you chose specific tools and parameters. In the future, you will thank yourself when you need to remember why you made certain decisions. Collaborators will also appreciate clear documentation when they use your workflows.
Most platforms automatically document tool versions and parameters. Save these records with your results. This documentation lets you or others reproduce your exact analysis later, which is essential for scientific validity.
Check Quality at Every Step
Do not skip quality control. Bad input data produces bad results, no matter how good your workflow is. Check that your sequencing quality is acceptable, your samples align well, and intermediate results look reasonable. Quality checks save time by catching problems before they cascade through your entire analysis.
Join Communities for Help
Every platform has user communities where people ask questions and share solutions. Join these forums, mailing lists, or chat groups. When you get stuck, search existing discussions or ask for help. Most community members remember being beginners and happily assist newcomers.
Real Examples of No-Code Workflows
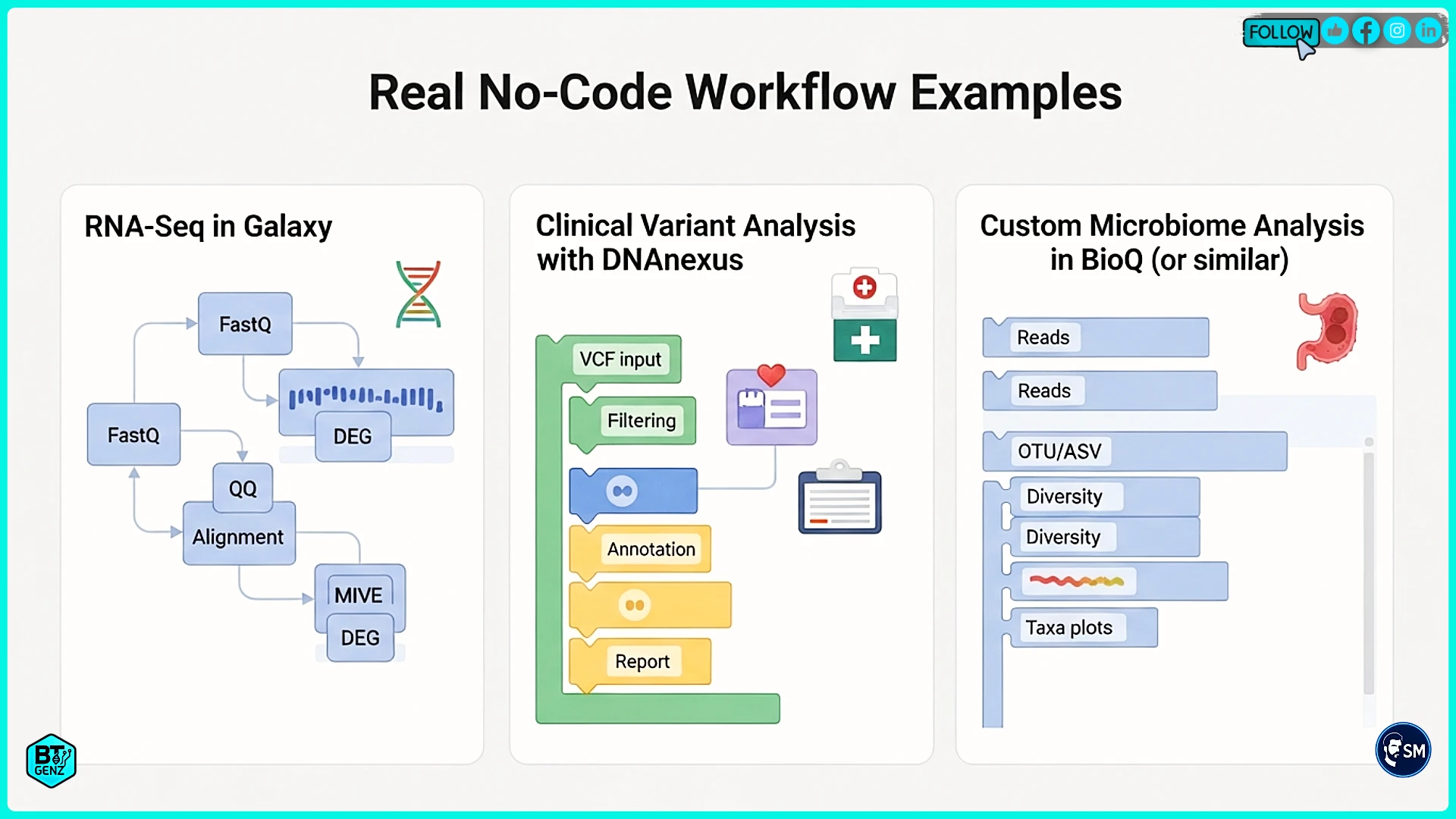
RNA-Seq Analysis in Galaxy
A cancer research lab needed to find genes with different expression between tumors and normal tissue. The biologists understood cancer biology but could not program. They used Galaxy with a published RNA-seq workflow template.
The workflow processed raw sequencing files through quality checking, alignment to the human genome, counting how many reads hit each gene, and statistical analysis to find differences. Galaxy made each step visual and understandable. Results identified hundreds of differentially expressed genes that the team validated in the lab.
Clinical Variant Analysis with DNAnexus
A hospital laboratory analyzes patient genomes to diagnose genetic diseases. They needed a secure, compliant infrastructure meeting healthcare privacy rules. DNAnexus provided workflows for processing patient samples, finding genetic variants, and annotating which variants might cause disease.
Clinical staff without programming backgrounds run standardized workflows on each patient. Automated quality checks flag samples needing repeat testing. AI prioritization highlights which variants doctors should examine closely. This system reduced diagnosis time while improving accuracy.
Custom Microbiome Analysis with Bionl
A researcher studying soil bacteria needed a custom analysis not available in standard workflows. They described their needs to Bionl in plain English: "Analyze metagenomic data to find bacterial species and compare diversity across sample types."
Bionl's AI built a complete workflow with appropriate tools for taxonomic classification and diversity analysis. When the researcher needed custom diversity calculations, the AI generated Python code implementing exactly what they needed. This hybrid approach delivered flexibility without requiring programming expertise.
Common Mistakes to Avoid
Wrong Data Formats
Data format problems cause most workflow failures. Always verify your files match the format the tools expect. Check file extensions, compression types, and whether index files are needed. Small format mismatches make tools fail with confusing errors.
Skipping Quality Control
Never skip quality checks on your input data. Bad data produces bad results even with perfect workflows. Always examine sequencing quality, check for contamination, and verify your samples are what you think they are before running major analyses.
Ignoring Resource Needs
Large datasets need substantial computer memory and storage. Workflows fail if you do not allocate enough resources. Start with recommended settings and increase if jobs fail. Some platforms automatically suggest appropriate resource levels based on your data size.
Not Validating Results
Always check that results make biological sense. Compare your findings to published studies of similar data. Use positive controls with known characteristics when possible. Surprising results might be real discoveries or might indicate workflow problems. Validation helps you tell the difference.
Getting Professional Help
While no-code platforms make bioinformatics accessible, complex projects sometimes benefit from expert guidance. Consider professional help when you feel stuck choosing the right approach, need help designing analysis strategies for unusual experiments, or want to establish best practices for your team.
Services like Visibility Accelerator provide specialized support for bioinformatics professionals. Whether you need help building specific workflows, optimizing existing analyses, or training your team on best practices, expert consultation can accelerate your progress significantly.
Professional services are particularly valuable when you are establishing computational capabilities for the first time. Getting expert help upfront prevents months of trial and error while establishing solid foundations for future work.
Next Steps and Resources
Start Practicing Today
Choose one platform from this guide and create a free account. Galaxy and GenePattern are excellent starting points because they cost nothing and have extensive tutorials. Complete a basic tutorial using their example data to learn the interface.
Next, try analyzing a small sample of your own data. Start with standard workflows before attempting custom analyses. Do not worry about making mistakes. Everyone struggles initially, and practice is how you learn. The bioinformatics community is welcoming and helpful when you ask questions.
Learning Resources
Galaxy Project offers free training materials covering everything from basics to advanced topics at their training website. Video tutorials walk through common analyses step by step using real datasets. The Galaxy Help Forum answers thousands of questions from users worldwide.
GenePattern provides detailed tutorials for its platform with example datasets. The Broad Institute also hosts webinars teaching specific analysis types. Most platforms maintain documentation wikis explaining every tool and feature in detail.
Join Communities
Connect with other bioinformatics learners and experts through online communities. Biostars is a question and answer site for bioinformatics where people share solutions to common problems. Reddit has active bioinformatics communities discussing tools and techniques. Many platforms also maintain Slack or Discord channels for real-time help.
The Future of No-Code Bioinformatics
Artificial intelligence will make no-code platforms even more powerful over the coming years. AI will not just help build workflows but will actively participate in analysis design. Future platforms will suggest experiments based on your results, generate hypotheses from data patterns, and recommend follow-up analyses.
Technologies currently requiring expert knowledge, like single-cell analysis, spatial transcriptomics, and multi-omics integration, will become accessible through simple interfaces. As methods mature, analyses once limited to specialized labs will spread to any researcher with questions to answer.
Cloud computing will continue making powerful analysis available to everyone regardless of local computer capabilities. The distinction between desktop and supercomputer will blur as platforms automatically scale resources based on what each analysis needs.
Conclusion
No-code AI workflows have opened bioinformatics to everyone with biological questions to answer. You no longer need programming expertise to conduct sophisticated computational analyses. Visual interfaces, AI assistance, and supportive communities make analysis accessible regardless of your technical background.
Start today by choosing one platform and completing a tutorial. Try analyzing a small sample of your own data. Join communities where you can ask questions and learn from experienced users. With practice, you will soon conduct analyses that seemed impossible just months ago.
Your biological expertise, combined with accessible computational tools, positions you to make real discoveries. The questions you can answer are limited only by your curiosity and data availability, not by programming ability. The future of biological research is computational, and no-code platforms ensure that future includes all researchers.
For more guidance on building your bioinformatics career, read our articles on how to apply for a Bioinformatics PhD, showcasing projects globally, and using LinkedIn for visibility. These resources provide complete strategies for advancing in computational biology.
References
Frequently Asked Questions
Founder of BTGenZ. Passionate about simplifying biotechnology for the next generation and bridging the information gap for aspiring biotechnologists in India.

Related Reads
Engage with Our Community
Join the conversation and share your thoughts with the BTGenZ community!
Connect on LinkedInLoading commenting section...
Comments Section
No approved comments yet. Be the first to leave a comment!